В современном мире, где технологии развиваются с невероятной скоростью, многие из нас сталкиваются с необходимостью обновления своей...
Покупка земельного участка — это серьезное решение и долгосрочная инвестиция, поэтому перед тем, как принять окончательное решение,...
Приобретение земельного участка может быть ключевым шагом в достижении желаемой свободы и комфорта. Будь то для застройки...
Поселки Московской области Московская область – один из крупнейших субъектов Российской Федерации, расположенный в центральной части страны....
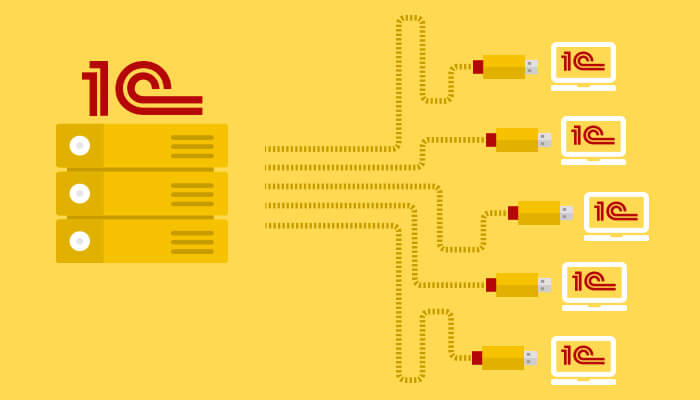
Аренда сервера для 1С: что это такое? Аренда сервера для 1С предоставляет компаниям возможность воспользоваться выделенным серверным...
Понадобилась дополнительная финансовая поддержка для развития своего бизнеса? Срочный займ может стать отличным решением для вас. В...
Советы по приготовлению раствора для мыльных пузырей в домашних условиях — легкий и эффективный рецепт
1 мин чтения
Мыльные пузыри — это удивительное развлечение, которое популярно не только среди детей, но и у взрослых. Прекрасно...
Минтай — это одна из самых распространенных рыб в наших водах. Он отличается нежным мясом и неповторимым...
Цветная капуста — это овощ с нежным вкусом и великолепным ароматом, который можно использовать для создания самых...
Томатный соус из томатной пасты — это прекрасное блюдо, которое может использоваться во множестве рецептов. Он отлично...